Hi,
Thanks for reading this letter. I got the following question and wish
someone could help me.
Given two graphs A and B, the vertices in these two network are one to one
correlated.
For example:
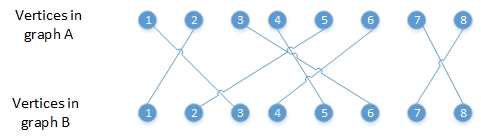
When generate subgraph A1 from graph A which contain vertices,
like subrootA <- c(2, 4, 6, 7),
the corresponding subgraph B1 in B should contains the vertices,
like subrootB <- c(1, 5, 4, 8).

Using
A1
<- induced.subgraph(A, subrootA, impl="create_from_scratch")
B1
<- induced.subgraph(B, subrootB, impl="create_from_scratch")
we should get
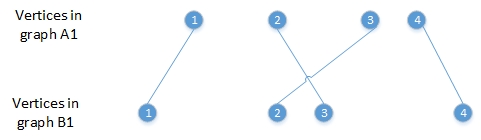
I used a vertice to encode the one-to-one corresponding relationship: AtoB
<- c(3, 1, 6, 5, 2, 4, 8, 7).
When using induced.subgraph, the vertex ids changes.
The vertice to encode the relationship between graphA1 and graphB1 should
be AtoB1 == c(1, 3, 2, 4)
I used the following code to solove this:
AtoB <- c(3, 1, 6, 5, 2, 4, 8, 7)
subrootA <- c(2, 4, 6, 7)
temp <- AtoB[subrootA]
N <- length(temp)
AtoB1<- rep(0, N)
M <- max(temp)
for(i in 1 : N)
{
min <- M
i_min <- 0
for(j in 1 : N)
{
if(min >= temp[j])
{
min <- temp[j]
i_min <-j
}
}
AtoB1[i_min] <- i
temp[i_min] <- M+ 1
}
> AtoB1
[1] 1 3 2 4
However, the time complexity is O(N2).
When N is larger than 1000000, it takes too much time.
Is there any faster way to fix it?
It is a long question, I wish that I had put it clearly :)
If not, please let me know. Thank you.
best
Xueming
address@hidden